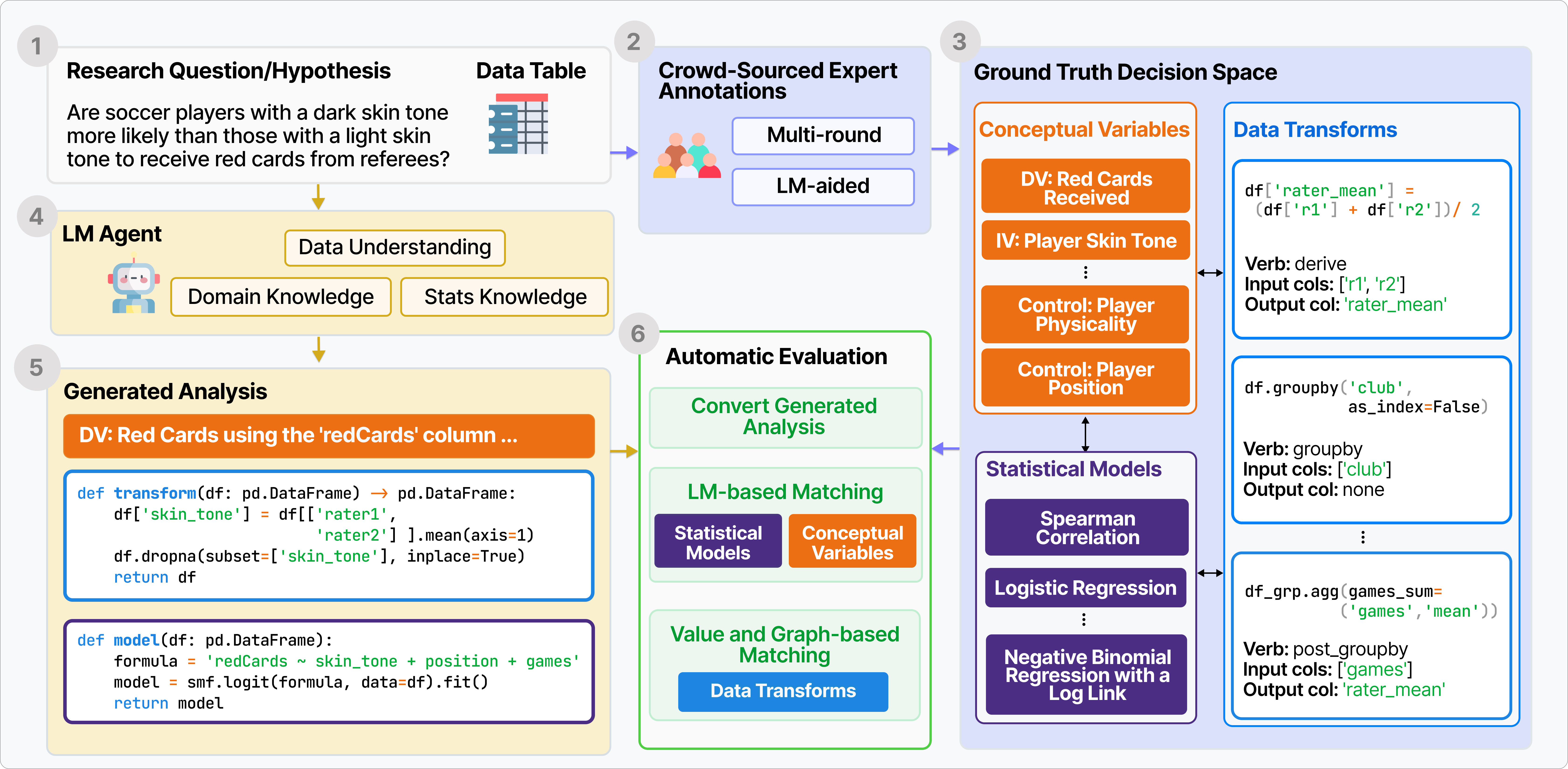
 BLADE automatically evaluates on the intermediate decisions (3)
to open-ended data-driven research questions (1). We gathered research questions and datasets
from existing research papers, crowd-sourced analysis studies and statistic textbooks as well as
analyses
from expert annotators (boxes 1-2-3).
Given a research question and dataset, LM agents generate a full
analysis containing the relevant conceptual variables, a data transform function, and a statistical
modeling function (boxes 1-4-5).
BLADE automatically evaluates on the intermediate decisions (3)
to open-ended data-driven research questions (1). We gathered research questions and datasets
from existing research papers, crowd-sourced analysis studies and statistic textbooks as well as
analyses
from expert annotators (boxes 1-2-3).
Given a research question and dataset, LM agents generate a full
analysis containing the relevant conceptual variables, a data transform function, and a statistical
modeling function (boxes 1-4-5).
 BLADE automatically evaluates this against the ground truth.
BLADE automatically evaluates this against the ground truth.
 BLADE consists of real-world open-ended research questions. These
questions
naturally elicit justifiable alternative decisions, from data preprocessing to statistical
modeling. We
aim to capture this flexibility in our ground truth annoation process.
BLADE consists of real-world open-ended research questions. These
questions
naturally elicit justifiable alternative decisions, from data preprocessing to statistical
modeling. We
aim to capture this flexibility in our ground truth annoation process.
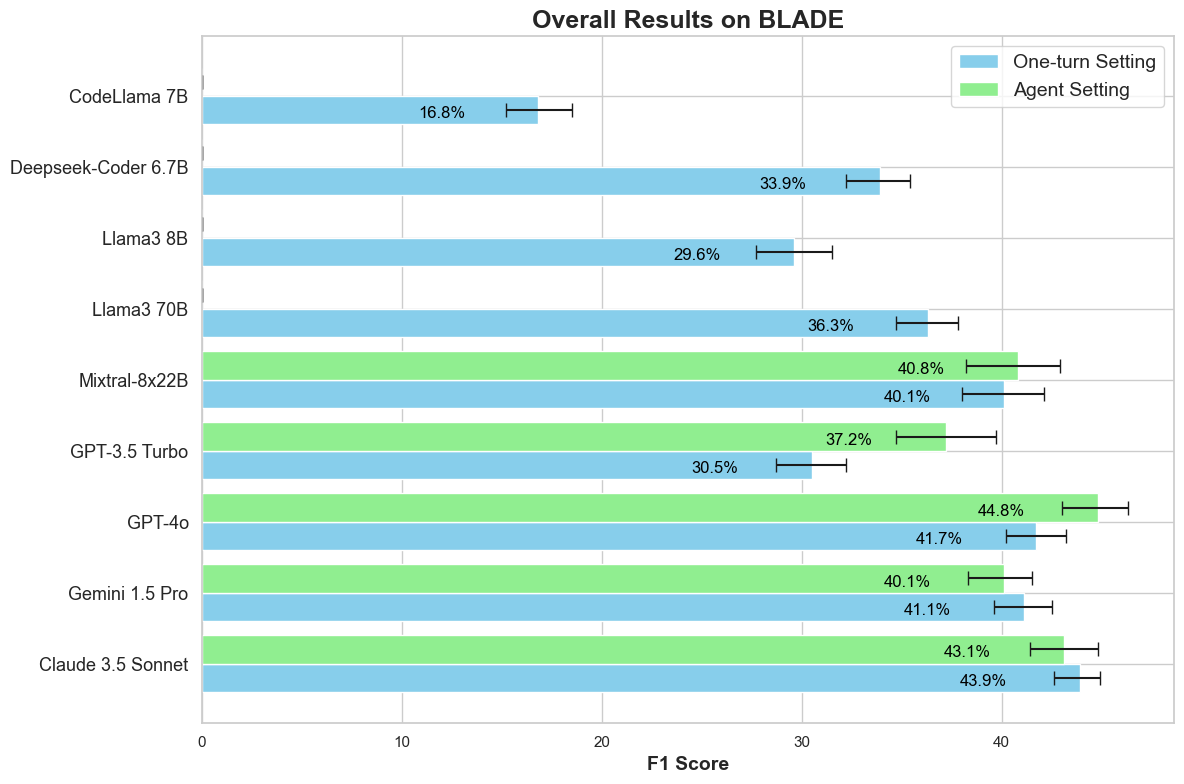
We evaluate open source and close source LMs on  BLADE in both a one turn direct prompting setting and a ReAct
agent
setting in
which the agent can interact with a sandbox notebook environment. The best setting achieves a F1-score
of 44.8% leaving lots of room for improvement.
BLADE in both a one turn direct prompting setting and a ReAct
agent
setting in
which the agent can interact with a sandbox notebook environment. The best setting achieves a F1-score
of 44.8% leaving lots of room for improvement.
Data-driven scientific discovery requires the iterative integration of scientific domain knowledge, statistical expertise, and an understanding of data semantics to make nuanced analytical decisions, such as which variables, transformations, and statistical models to consider. LM-based agents equipped with planning, memory, and code execution capabilities have the potential to support data-driven science; however, evaluating these agents on open-ended tasks is challenging due to multiple valid approaches, partially correct steps, and different ways to express the same decisions.
To address these challenges, we present  BLADE, a benchmark designed to
automatically evaluate agents’ multifaceted approaches to open-ended
research questions.
BLADE, a benchmark designed to
automatically evaluate agents’ multifaceted approaches to open-ended
research questions.  BLADE consists of 12 datasets and research questions
drawn from existing scientific literature, with ground truth collected
from independent analyses by expert data scientists and researchers. To
facilitate the automatic evaluation of agent responses, we developed
computational methods that match different representations of analyses
to this ground truth. Although language models possess considerable world
knowledge, our evaluation shows they are often limited to basic analyses;
in contrast, agents capable of interacting with the underlying
data demonstrate improved, yet still non-optimal, diversity
in their analytical decision-making. Our work enables the evaluation
of agents in the context of data-driven science and provides researchers
with deeper insights into the analytical approaches employed by these agents.
BLADE consists of 12 datasets and research questions
drawn from existing scientific literature, with ground truth collected
from independent analyses by expert data scientists and researchers. To
facilitate the automatic evaluation of agent responses, we developed
computational methods that match different representations of analyses
to this ground truth. Although language models possess considerable world
knowledge, our evaluation shows they are often limited to basic analyses;
in contrast, agents capable of interacting with the underlying
data demonstrate improved, yet still non-optimal, diversity
in their analytical decision-making. Our work enables the evaluation
of agents in the context of data-driven science and provides researchers
with deeper insights into the analytical approaches employed by these agents.
 BLADE Dataset
BLADE Dataset
 BLADE is an expert annotated benchmark studying agent performance on
open-ended data-driven research questions (there is no single answer result).
BLADE is an expert annotated benchmark studying agent performance on
open-ended data-driven research questions (there is no single answer result).
 BLADE evaluates how well agents can understand data, integrate this with
external scientific
domain knowledge, and execute an analysis. In particular,
BLADE evaluates how well agents can understand data, integrate this with
external scientific
domain knowledge, and execute an analysis. In particular,  BLADE focuses on the following tasks, :
BLADE focuses on the following tasks, :
We source research
questions from existing crowd-sourced
analysis studies and statistics textbooks. We follow a rigourous multi-stage annotation procedure to
ensure annotation
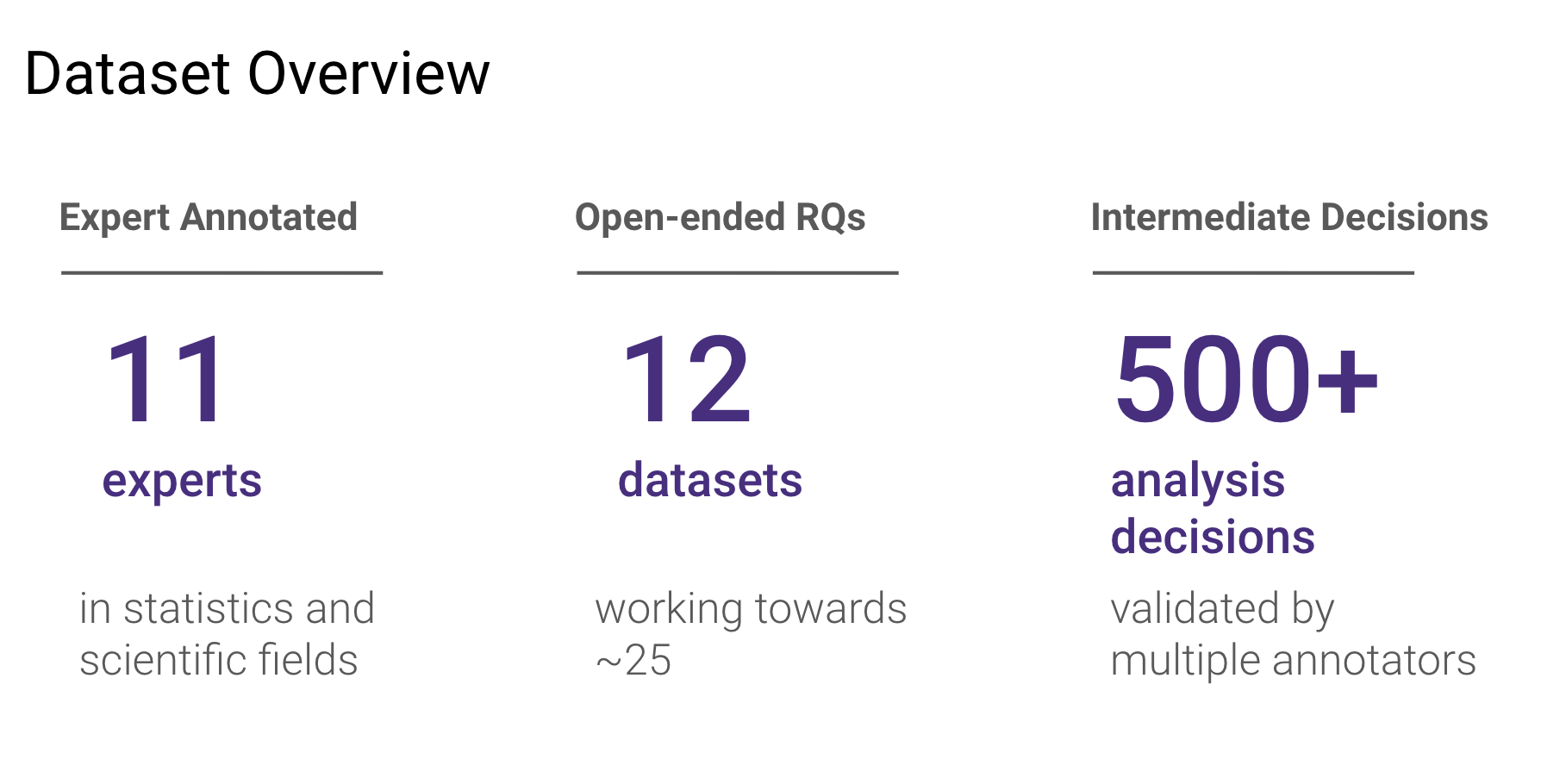
quality.  BLADE currently consists of 12 research questions and datasets
encompassing over 500
analysis decisions.
BLADE currently consists of 12 research questions and datasets
encompassing over 500
analysis decisions.
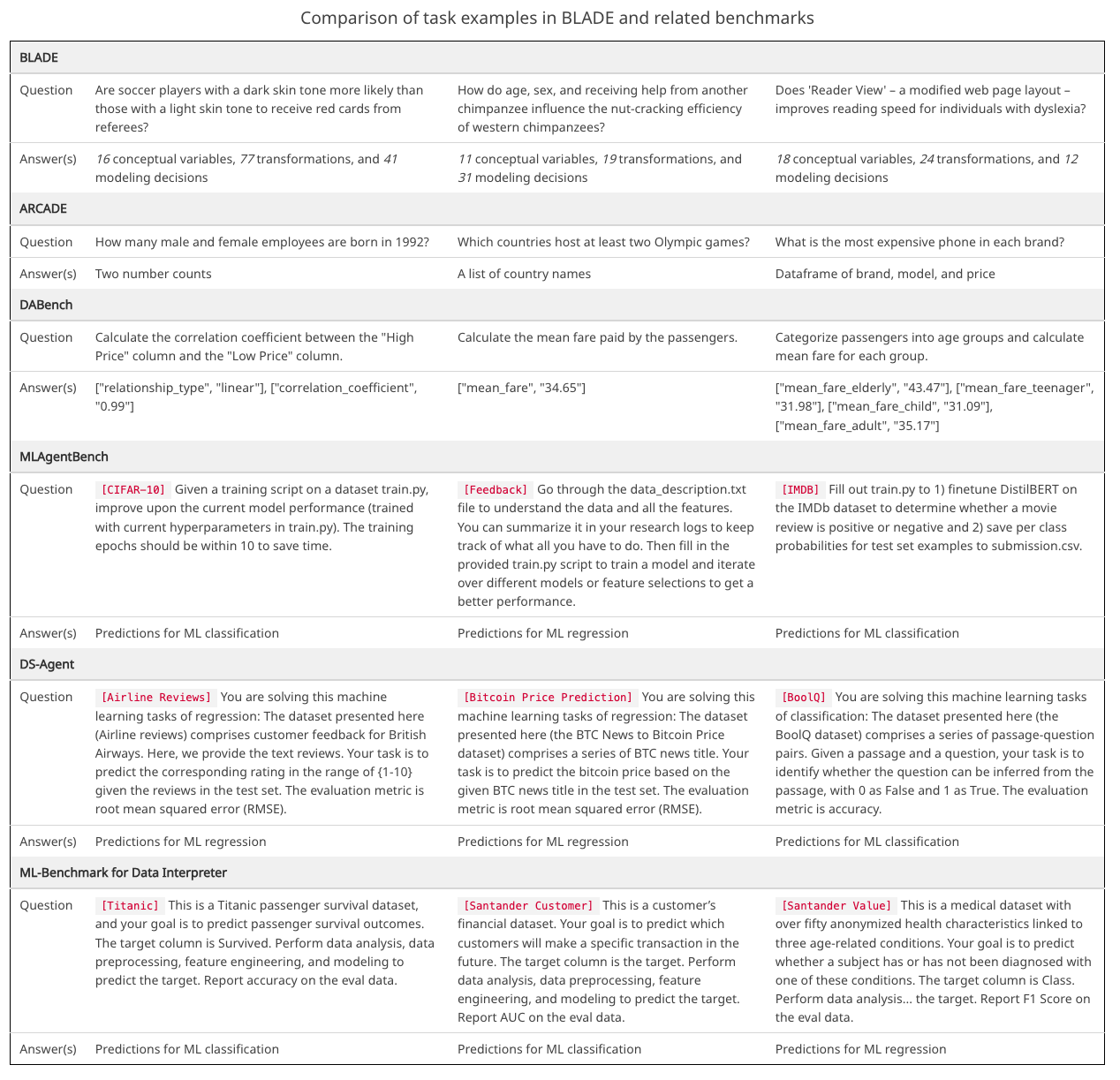
Examples of tasks in our work and related benchmarks.
 BLADE prioritizes open-ended research questions and considers the
flexibility of analysis approaches in its evaluation.
BLADE prioritizes open-ended research questions and considers the
flexibility of analysis approaches in its evaluation.
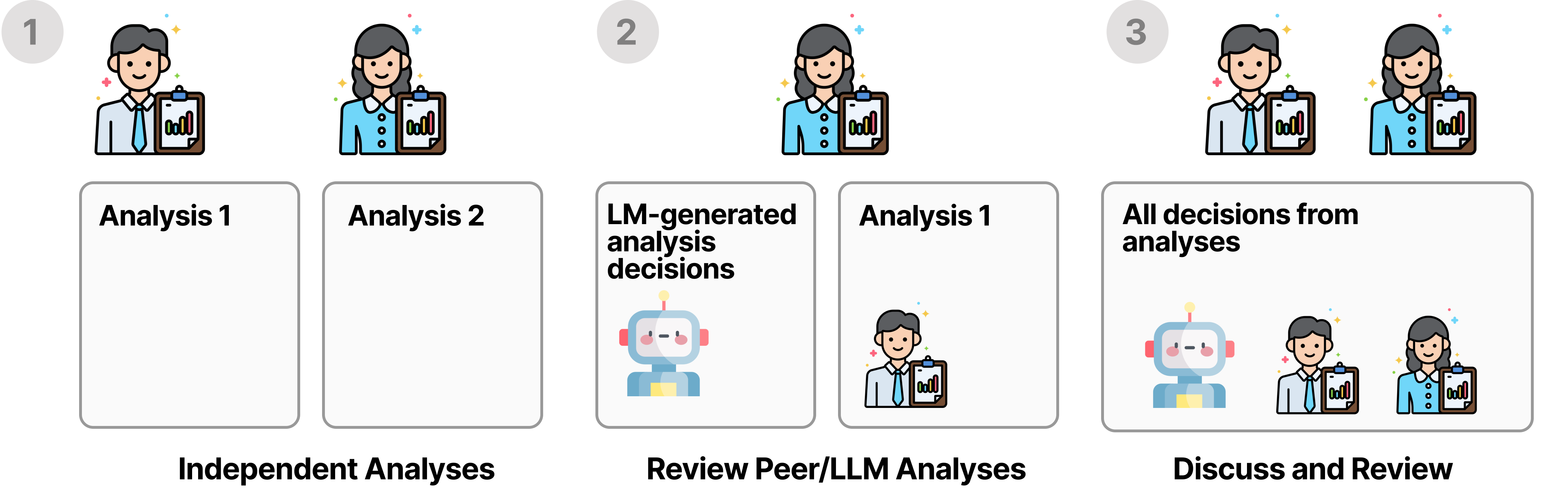
 BLADE follows a multi-stage annotation procedure to ensure
ground-truth
annotation quality.
First, annotators independetly conduct their own analysis given a research question and dataset and
report their analysis decisions (1).
Next, each annotator reviews the analysis decisions of the other annotators and those generated by a
LM to validate each other's approach and also consider new potentially overlooked approaches (2).
Finally, annotators come together, discuss their analysis decisions and reach a consensus on the
ground-truth analysis decisions (3).
BLADE follows a multi-stage annotation procedure to ensure
ground-truth
annotation quality.
First, annotators independetly conduct their own analysis given a research question and dataset and
report their analysis decisions (1).
Next, each annotator reviews the analysis decisions of the other annotators and those generated by a
LM to validate each other's approach and also consider new potentially overlooked approaches (2).
Finally, annotators come together, discuss their analysis decisions and reach a consensus on the
ground-truth analysis decisions (3).
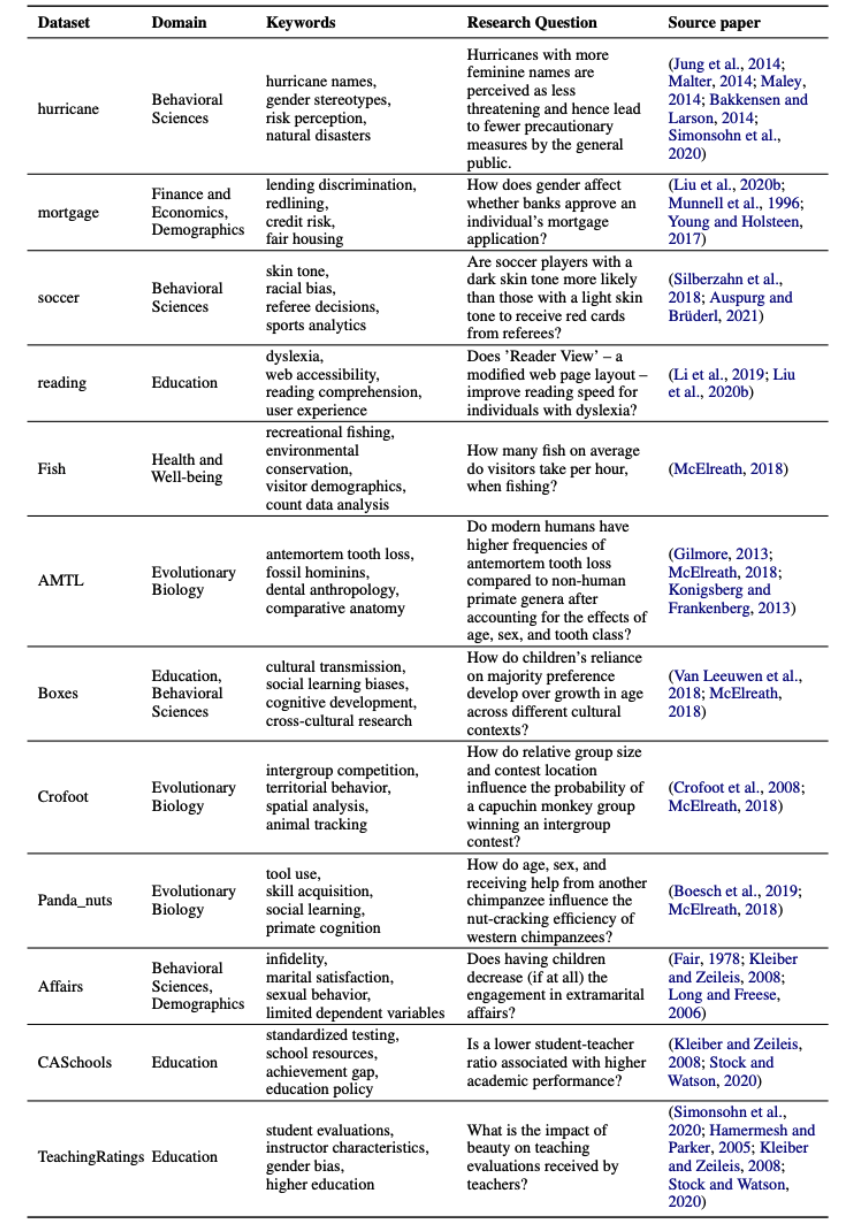
Summary of the 12 different source datasets in  BLADE.
BLADE.
Given a research question and data table, the agent's submission of an analysis to BLADE is a JSON file containing the following elements:

We evaluate open source and close source LMs on  BLADE in both a one turn direct prompting setting and a ReAct
agent
setting in
which the agent can interact with a sandbox computational notebook environment.
BLADE in both a one turn direct prompting setting and a ReAct
agent
setting in
which the agent can interact with a sandbox computational notebook environment.

 BLADE indicating that there is still a lot of room for improvement.
Generally, the ReAct agent outperforms the one turn direct prompting setting.
BLADE indicating that there is still a lot of room for improvement.
Generally, the ReAct agent outperforms the one turn direct prompting setting.
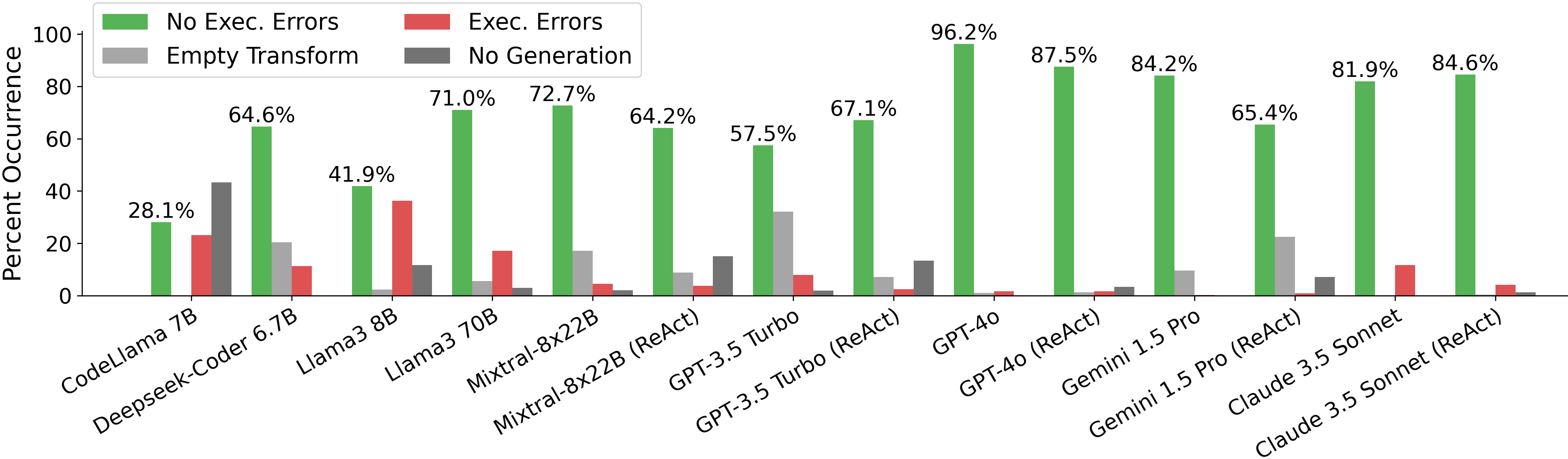
For generating an analysis, we find that most large LMs can generate a non-empty executable analysis over 60% of the time, with GPT-4o being the best at 96%. Among the open-source models, Mixtral-8x22b performs best, generating an executable analysis 73% of the time and DeepSeek-Coder also does surprisingly well at 65%.
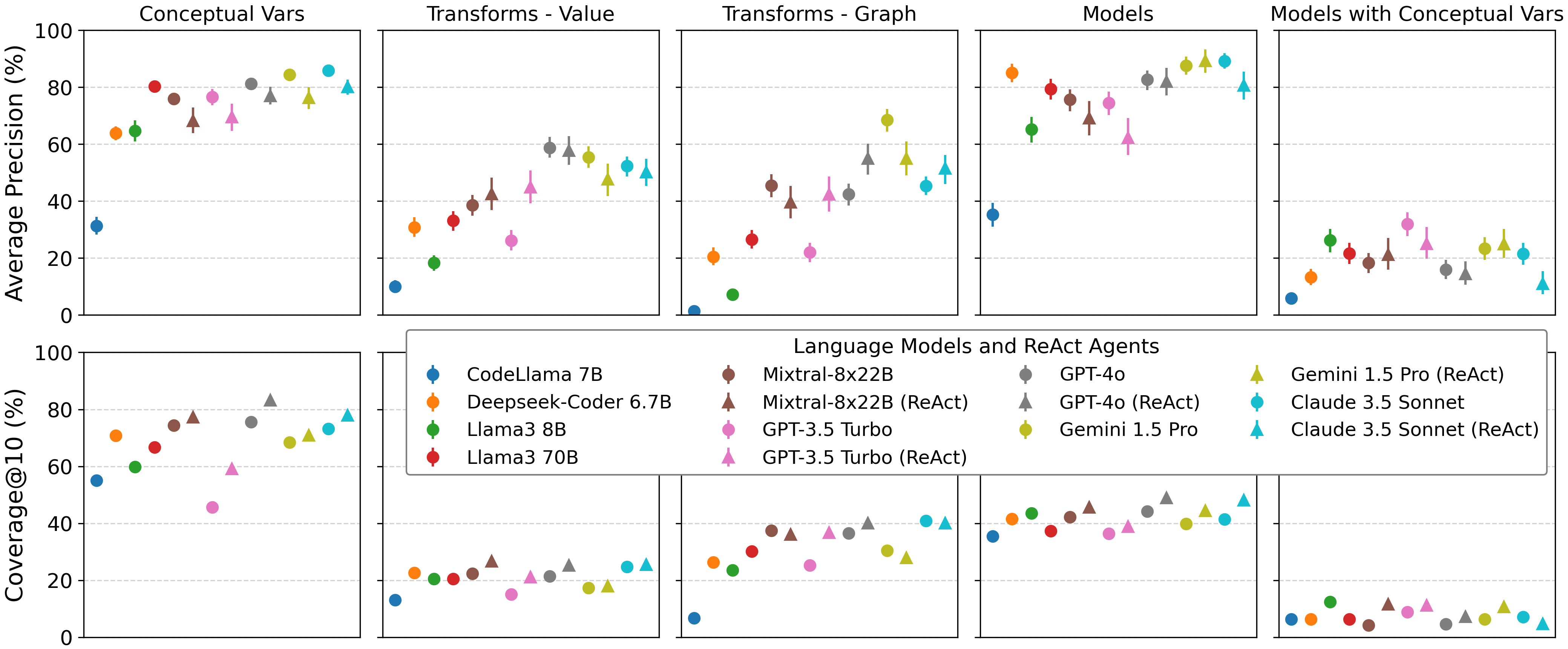
Average precision (top row) and coverage@10 (bottom row) percentages averaged across datasets in  BLADE. All runs were included in the results.
Run errors default to a hit rate of 0 and are counted in the coverage calculation (i.e., treated as a
run
that generated nothing).
Error bars represent bootstrapped 95% confidence intervals.
BLADE. All runs were included in the results.
Run errors default to a hit rate of 0 and are counted in the coverage calculation (i.e., treated as a
run
that generated nothing).
Error bars represent bootstrapped 95% confidence intervals.
We also observe low coverage of the ground truth examples, especially with respect to data transformations and specific model specifications. Through qualitatively reviewing a random sample of LM-generated analyses, we find that LMs often perform basic analysis that can yield decent precision (i.e., matching basic decisions) but poor coverage across runs. However, comparing the one-turn and agent settings, LMs consistently had higher coverage when allowed to iteratively explore data. Moreover, ReAct agents perform best overall on coverage for data transformations and statistical modeling, which require a more detailed understanding of data semantics.
See the examples of generated analyses using GPT-4o (one of the best performing LMs) via direct prompting and the ReAct framework below.
GPT-4o one turn direct prompting examples:
# ======== TRANSFORM CODE ========
def transform(df: pd.DataFrame) -> pd.DataFrame:
# Filter out rows with missing values in rater1 and rater2
df = df.dropna(subset=['rater1', 'rater2'])
# Calculate the average skin tone from rater1 and rater2
df['AvgSkinTone'] = df[['rater1', 'rater2']].mean(axis=1)
# Return the transformed dataframe
return df
# ======== MODEL CODE ========
def model(df: pd.DataFrame) -> Any:
# Prepare the independent variables and control variables
X = df[['AvgSkinTone', 'games', 'yellowCards', 'meanIAT', 'meanExp']]
y = df['redCards']
# Add a constant to the model (intercept)
X = sm.add_constant(X)
# Perform the linear regression
model = sm.OLS(y, X).fit()
# Return the results of the model
return model.summary() Ex. 1) answering the question Are soccer players with a dark skin tone more likely than those with a light skin tone to receive red cards from referees?
# ======== TRANSFORM CODE ========
def transform(df: pd.DataFrame) -> pd.DataFrame:
# Replace missing skin tone ratings with the mean rating
df['rater1'] = df['rater1'].fillna(df['rater1'].mean())
df['rater2'] = df['rater2'].fillna(df['rater2'].mean())
# Calculate the average skin tone rating
df['SkinTone'] = (df['rater1'] + df['rater2']) / 2
# Return the transformed dataframe with only necessary columns
return df[['SkinTone', 'redCards', 'games', 'meanIAT', 'meanExp']]
# ======== MODEL CODE ========
def model(df: pd.DataFrame) -> Any:
# Fit the regression model
X = df[['SkinTone', 'games', 'meanIAT', 'meanExp']]
y = df['redCards']
X = sm.add_constant(X)
model = sm.OLS(y, X).fit()
# Return the model results
return model.summary()Ex. 2) answering the question Are soccer players with a dark skin tone more likely than those with a light skin tone to receive red cards from referees?
# ======== TRANSFORM CODE ========
def transform(df: pd.DataFrame) -> pd.DataFrame:
# Replace zero rating with NaN for rater columns indicating missing ratings
df['rater1'].replace(0, np.nan, inplace=True)
df['rater2'].replace(0, np.nan, inplace=True)
# Drop rows where both 'rater1' and 'rater2' are NaN
df.dropna(subset=['rater1', 'rater2'], how='all', inplace=True)
# Calculate the average skin tone rating
df['avgSkinTone'] = df[['rater1', 'rater2']].mean(axis=1)
# One-hot encode the position column
df = pd.get_dummies(df, columns=['position'], drop_first=True)
return df
# ======== MODEL CODE ========
def model(df: pd.DataFrame) -> Any:
# Define the independent variables including controls
indep_vars = ['avgSkinTone', 'games', 'meanIAT', 'meanExp'] + [col for col in df.columns if col.startswith('position_')]
X = df[indep_vars]
# Define the dependent variable
y = df['redCards']
# Add a constant to the model (intercept)
X = sm.add_constant(X)
# Fit the OLS model
model = sm.OLS(y, X)
results = model.fit()
# Return the model summary
return results.summary()Ex. 3) answering the question Are soccer players with a dark skin tone more likely than those with a light skin tone to receive red cards from referees?
# ======== TRANSFORM CODE ========
def transform(df: pd.DataFrame) -> pd.DataFrame:
# Drop rows with missing values in the essential columns
df = df.dropna(subset=['rater1', 'rater2', 'redCards', 'meanIAT', 'meanExp', 'games'])
# Calculate the average skin tone based on the two raters
df['AvgSkinTone'] = df[['rater1', 'rater2']].mean(axis=1)
# Return the transformed dataframe ready for modeling
return df
# ======== MODEL CODE ========
def model(df: pd.DataFrame) -> Any:
# Define the independent variables (including control variables)
X = df[['AvgSkinTone', 'meanIAT', 'meanExp', 'games']]
# Add a constant to the model (intercept)
X = sm.add_constant(X)
# Define the dependent variable
y = df['redCards']
# Fit an OLS regression model
model = sm.OLS(y, X).fit()
# Return the summary of the model
return model.summary()
Ex. 4) answering the question Are soccer players with a dark skin tone more likely than those with a light skin tone to receive red cards from referees?
# ======== TRANSFORM CODE ========
def transform(df: pd.DataFrame) -> pd.DataFrame:
# Replace zero rating with NaN for rater columns indicating missing ratings
df['rater1'].replace(0, np.nan, inplace=True)
df['rater2'].replace(0, np.nan, inplace=True)
# Drop rows where both 'rater1' and 'rater2' are NaN
df.dropna(subset=['rater1', 'rater2'], how='all', inplace=True)
# Calculate the average skin tone rating
df['avgSkinTone'] = df[['rater1', 'rater2']].mean(axis=1)
# One-hot encode the position column
df = pd.get_dummies(df, columns=['position'], drop_first=True)
return df
# ======== MODEL CODE ========
def model(df: pd.DataFrame) -> Any:
# Define the independent variables including controls
indep_vars = ['avgSkinTone', 'games', 'meanIAT', 'meanExp'] + [col for col in df.columns if col.startswith('position_')]
X = df[indep_vars]
# Define the dependent variable
y = df['redCards']
# Add a constant to the model (intercept)
X = sm.add_constant(X)
# Fit the OLS model
model = sm.OLS(y, X)
results = model.fit()
# Return the model summary
return results.summary()Ex. 5) answering the question Are soccer players with a dark skin tone more likely than those with a light skin tone to receive red cards from referees?
GPT-4o ReAct agent examples:
# ======== TRANSFORM CODE ========
def transform(df: pd.DataFrame) -> pd.DataFrame:
# Drop rows with missing values in critical columns
df_clean = df.dropna(subset=['rater1', 'rater2', 'position', 'meanIAT', 'meanExp'])
# Create a new column for the average skin tone rating
df_clean['avgSkinTone'] = df_clean[['rater1', 'rater2']].mean(axis=1)
return df_clean
# ======== MODEL CODE ========
import statsmodels.api as sm
def model(df: pd.DataFrame) -> sm.GLM:
# Define the control variables
control_vars = ['games', 'meanIAT', 'meanExp']
# Create dummy variables for the categorical variable 'position'
df = pd.get_dummies(df, columns=['position'], drop_first=True)
# Define the dependent variable
y = df['redCards']
# Define the independent variables including dummy variables for position
X = df[['avgSkinTone'] + control_vars + [col for col in df.columns if col.startswith('position_')]]
# Add a constant term to the independent variables
X = sm.add_constant(X)
# Fit a Poisson regression model
poisson_model = sm.GLM(y, X, family=sm.families.Poisson()).fit()
return poisson_model
Ex. 1) answering the question Are soccer players with a dark skin tone more likely than those with a light skin tone to receive red cards from referees?
# ======== TRANSFORM CODE ========
def transform(df: pd.DataFrame) -> pd.DataFrame:
# Drop rows with missing 'skinTone'
df = df.dropna(subset=['skinTone'])
# Fill missing 'position' values with the most frequent value
most_frequent_position = df['position'].mode()[0]
df['position'] = df['position'].fillna(most_frequent_position)
# Create composite skin tone score
df['skinTone'] = df[['rater1', 'rater2']].mean(axis=1)
# Generate binary variable for skin tone
median_skin_tone = df['skinTone'].median()
df['skinToneCategory'] = df['skinTone'].apply(lambda x: 'Dark' if x > median_skin_tone else 'Light')
# Convert Red Cards to Binary
df['RedCardReceived'] = df['redCards'].apply(lambda x: 1 if x > 0 else 0)
return df
# ======== MODEL CODE ========
def model(df: pd.DataFrame) -> Any:
import statsmodels.api as sm
import statsmodels.formula.api as smf
# Ensure categorical variables are treated as such
df['position'] = df['position'].astype('category')
df['refCountry'] = df['refCountry'].astype('category')
df['skinToneCategory'] = df['skinToneCategory'].astype('category')
# Simplify model by focusing on key variables
logit_model = smf.logit('RedCardReceived ~ skinToneCategory + games + C(refCountry)', data=df).fit()
return logit_model.summary()Ex. 2) answering the question Are soccer players with a dark skin tone more likely than those with a light skin tone to receive red cards from referees?
# ======== TRANSFORM CODE ========
def transform(df: pd.DataFrame) -> pd.DataFrame:
# Calculate average skin tone rating
df['avgSkinTone'] = df[['rater1', 'rater2']].mean(axis=1)
# Filter to include only players with valid skin tone ratings
df_filtered = df.dropna(subset=['avgSkinTone'])
# Summarize data at the player level
df_player_level = df_filtered.groupby(['playerShort'], as_index=False).agg({
'avgSkinTone': 'mean',
'redCards': 'sum',
'games': 'sum',
'meanIAT': 'mean',
'meanExp': 'mean'
})
return df_player_level
# ======== MODEL CODE ========
def model(df: pd.DataFrame):
import statsmodels.api as sm
import statsmodels.formula.api as smf
# Fit a negative binomial regression model
model = smf.glm('redCards ~ avgSkinTone + games + meanIAT + meanExp',
data=df,
family=sm.families.NegativeBinomial()).fit()
return model.summary()Ex. 3) answering the question Are soccer players with a dark skin tone more likely than those with a light skin tone to receive red cards from referees?
# ======== TRANSFORM CODE ========
def transform(df: pd.DataFrame) -> pd.DataFrame:
# Drop rows with missing values in rater1 and rater2
df_cleaned = df.dropna(subset=['rater1', 'rater2'])
# Calculate average skin tone score
df_cleaned['avgSkinTone'] = df_cleaned[['rater1', 'rater2']].mean(axis=1)
# Select necessary columns
columns = ['avgSkinTone', 'redCards', 'games', 'victories', 'ties', 'defeats', 'position', 'meanIAT', 'meanExp']
df_final = df_cleaned[columns]
return df_final
# ======== MODEL CODE ========
def model(df: pd.DataFrame) -> Any:
import statsmodels.formula.api as smf
# Fit the regression model
model = smf.ols('redCards ~ avgSkinTone + games + victories + ties + defeats + C(position) + meanIAT + meanExp', data=df).fit()
# Return the regression results
return model.summary() Ex. 4) answering the question Are soccer players with a dark skin tone more likely than those with a light skin tone to receive red cards from referees?
# ======== TRANSFORM CODE ========
def transform(df: pd.DataFrame) -> pd.DataFrame:
# Calculate the average skin tone rating
df['avgSkinTone'] = df[['rater1', 'rater2']].mean(axis=1)
# Filter relevant columns
columns_to_keep = ['avgSkinTone', 'redCards', 'games', 'meanIAT', 'meanExp', 'position', 'goals', 'victories', 'ties', 'defeats']
df = df[columns_to_keep]
# Drop rows with missing values in the average skin tone
df = df.dropna(subset=['avgSkinTone'])
# One-hot encode the 'position' column
df = pd.get_dummies(df, columns=['position'], drop_first=True)
return df
# ======== MODEL CODE ========
def model(df: pd.DataFrame) -> Any:
import statsmodels.api as sm
# Define the dependent and independent variables
X = df.drop(columns=['redCards'])
y = df['redCards']
# Add a constant to the model
X = sm.add_constant(X)
# Fit the OLS model
ols_model = sm.OLS(y, X).fit()
return ols_model.summary()Ex. 5) answering the question Are soccer players with a dark skin tone more likely than those with a light skin tone to receive red cards from referees?
 BLADE
BLADE
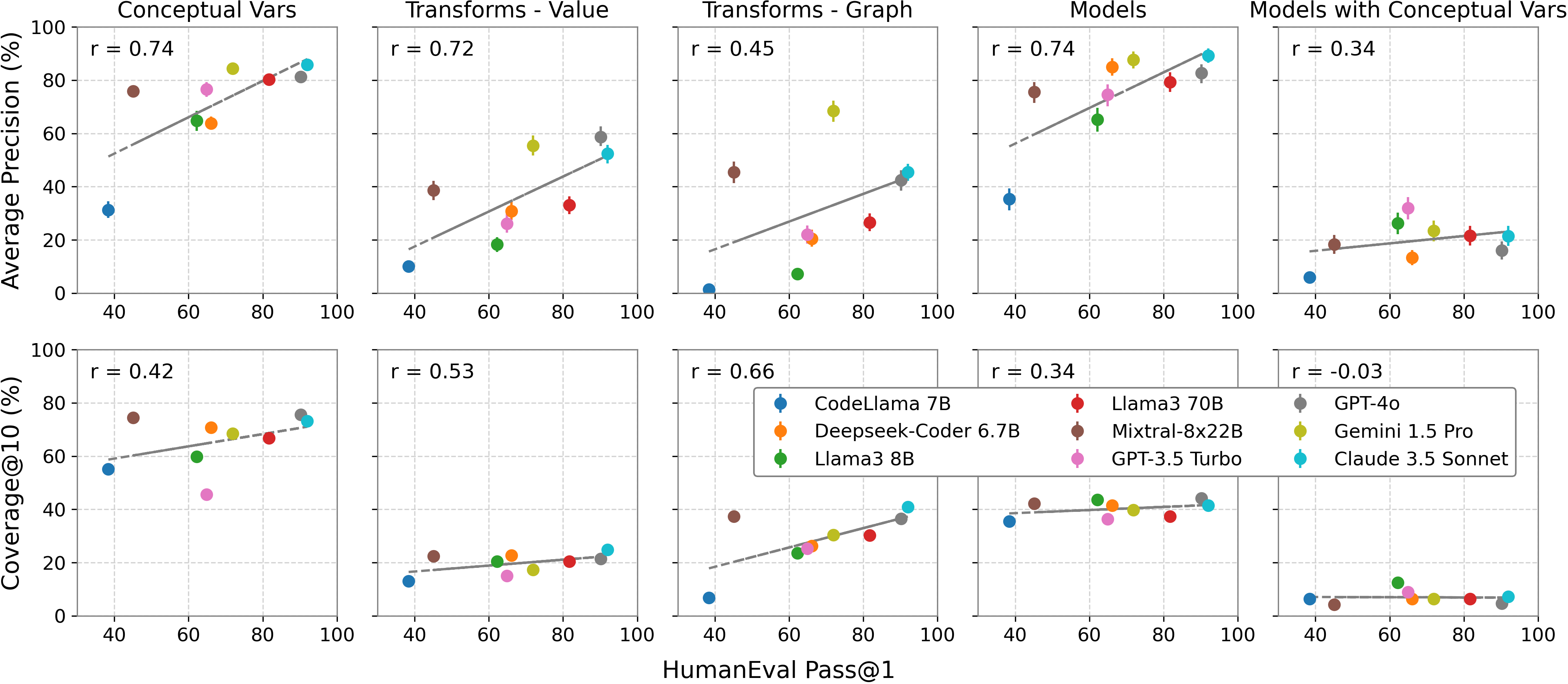
Performance vs. HumanEval Performance. We compare BLADE evaluation metrics against reported Pass@1 on HumanEval.
When comparing our results in analysis generation with those from the HumanEval coding benchmark, we fonud most metrics showed a positive correlation, indicating that higher HumanEval performance is broadly correlated with higher BLADE performance. However, coverage measures had a weaker correlation compared to precision. This suggests that while current training methods, such as Reinforcement Learning from Human Feedback (RLHF) and instruction tuning, optimize for one solution, they may struggle to generate diverse solutions.
@inproceedings{
gu2024blade,
title={BLADE: Benchmarking Language Model Agents for Data-Driven Science},
author={Ken Gu and Ruoxi Shang and Ruien Jiang and Keying Kuang and Richard-John Lin and Donghe Lyu and Yue Mao and Youran Pan and Teng Wu and Jiaqian Yu and Yikun Zhang and Tianmai M. Zhang and Lanyi Zhu and Mike A. Merrill and Jeffrey Heer and Tim Althoff},
booktitle={The 2024 Conference on Empirical Methods in Natural Language Processing},
year={2024},
}





